5 R basics
This tutorial is an introduction to R adapted from [1] with extra material from [2]. If you already have R experience, you might still want to browse this section in case you find something new.
Prerequisites should be completed before proceeding. After that, the tutorial should take no longer than 50 minutes.
5.1 R style guide
A style guide is about making your script readable. We ask you to observe a small set of guidelines that will help us help you when you have questions about your script.
Comments in R are denoted by a hash tag #.
- Everything to the right of the hash tag is ignored by R.
- Comments that describe “why” are generally more useful than comments that explain “how.”
Spaces around operators. Usewhitespacetoenhancereadability. Place spaces around operators (=, +, -, <-, etc.). Always put a space after a comma, but never before (just like in regular English).
# poor
height<-feet*12+inches
mean(x,na.rm=10)
# better
height <- (feet * 12) + inches
mean(x, na.rm = 10)Use vertical white space. Lack of vertical white space makes your script harder to read (like a story with no paragraphs).
# Poor, no paragraph breaks
library("data.table"); library("GDAdata")
speed_ski <- copy(SpeedSki)
setDT(speed_ski)
speed_ski <- speed_ski[, .(Event, Sex, Speed)]
setnames(speed_ski, old = c("Event", "Sex", "Speed"), new = c("event", "sex", "speed"))Instead, group chunks of code into paragraphs separated by blank lines to reveal the structure of the program. Comments at the start of a code chunk can explain your intent (like a topic sentence). Here we illustrate commenting on “why” rather than “how.”
# Better example with code in paragraphs
library("data.table")
library("GDAdata")
# Leave the original data unaltered before data.table conversion
speed_ski <- copy(SpeedSki)
setDT(speed_ski)
# Only three variables are required
speed_ski <- speed_ski[, .(Event, Sex, Speed)]
# Lowercase column names are our preferred style
setnames(speed_ski,
old = c("Event", "Sex", "Speed"),
new = c("event", "sex", "speed"))
# RDS format preserves factors
saveRDS(speed_ski, "data/speed_ski.rds")For more information of R scripting style generally see McConnell [3] and Wickham [4].
5.2 R follows a script
Use File > New File > R Script to create a new R script
- Name the script
01-R-basics.R. By using a number at the start of the file name, the files stay in order in your directory.
- Save it in the
scriptsdirectory. - Code chunks like the one below can be copied and pasted to your R script. Add a minimal header at the top of the script. Use
library()to load the packages we will be using.
# R basics
# name
# date
# packages
library("midfieldr")
library("data.table")After adding a code chunk to your script, run the script. Options for running a script:
- To run an entire script, select all lines with
ctrl Athen run the lines usingctrl Enter(for the Mac OS:cmd Aandcmd Return). - To run select lines, use the cursor to select the lines you want to run, then
ctrl Enter(for the Mac OS:cmd Return). - To run from the beginning to a line, place your cursor at the line, then
ctrl alt B(cmd option BMac OS)
Errors
If you get an error similar to:
Error in library("data.table") : there is no package called 'data.table'then the package needs to be installed. If you need a refresher on installing packages, see Install CRAN packages. Once the missing package is installed, you can rerun the script.
The following code chunk is optional for controlling the number of rows of a data frame that are printed to the Console screen.
# Optional code to control data.table printing
options(
datatable.print.nrows = 10,
datatable.print.topn = 5,
datatable.print.class = TRUE
)Healy [5] offers this advice for specific things to watch out for:
- Make sure parentheses are balanced—that every opening
(has a corresponding closing).
- Expect to make errors and don’t worry when that happens. You won’t break anything.
- Make sure you complete your expressions. If you see a
+in the Console instead of the usual prompt>, R thinks your expression is incomplete. For example, if you type the following and try to run it,
str(airqualitythe output in your Console reports:
#> str(airquality
#> + The plus sign indicates that the expression is incomplete…in this case a missing closing parenthesis. To recover, hit Esc or ctrl C. Then correct the code.
Interrupting R
If R seems to be taking too long to conclude a process, here are some things to try (depends on your OS) to recover.
Esc
ctrl C
- RStudio pulldown menu Session > Terminate R… > Yes
Guidelines
- As you work through the tutorial, type a line or chunk of code then File > Save and run the script.
- Confirm that your result matches the tutorial result.
- The exercises give you chance to devise your own examples and check them out. You learn by doing (but you knew that already)!
5.3 Everything in R has a name
In R, every object has a name.
- named entities, like
xory
- data you have loaded, like
my_data - functions you use, like
sin()
Some names are forbidden
- reserved words, like
TRUEorFALSE
- programming words, like
Inf,for,else, andfunction
- special entities, like
NAandNaN
Some names should not be used because they name commonly used functions
q()quitc()combine or concatenatemean()range()var()variance
Names in R are case-sensitive
my_dataandMy_Dataare different objects- We use the style of naming things in lower case with words separated by underscores (no spaces), e.g.,
speed_ski. The camel-case is also popular, e.g.,SpeedSkiorspeedSki. The choice is yours.
If you want to know if a name has already been used in a package you have loaded, go to the RStudio console, type a question mark followed by the name, e.g.,
# Type in the Console
? c()
? mean()If the name is in use, a help page appears in the RStudio Help pane.
5.4 Everything in R is an object
Origins of R objects
- Some objects are built in to R
- Some objects are loaded with packages
- Some objects are created by you
Type this line of code in your script, Save. c() is the function to combine or concatenate its elements to create a vector.
# Type in the R script
c(1, 2, 3, 1, 3, 25)Run the script and your Console should show [1] 1 2 3 1 3 25.
In these notes, when we show results printed in your Console, we preface the printout with #> (which does not appear on your screen) to distinguish the results from the script. For example, we show the line from above and its output like this:
c(1, 2, 3, 1, 3, 25) # <- typed in the script
#> [1] 1 2 3 1 3 25 # <- appears in the ConsoleThe [1] that leads the output line is a label identifying the index of the element that starts that line. More on that in a little while.
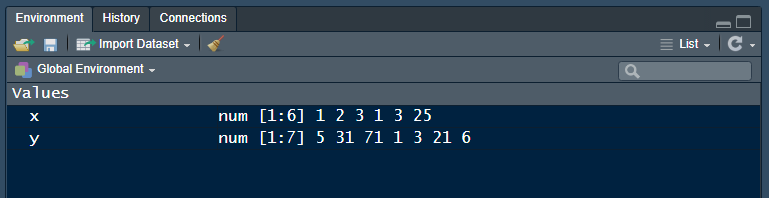
You create objects my assigning them names using the <- operator. The keyboard shortcut for the assignment operator is alt\(-\) , i.e., the ALT key plus the hyphen key. (Mac OS option\(-\))
# Practice assigning an object to a name
x <- c(1, 2, 3, 1, 3, 25)
y <- c(5, 31, 71, 1, 3, 21, 6)To see the result in the Console, type the object name in the script, Save, and run. (Remember, type the line of code but not the line prefaced by #>—that’s the output line so you can check your results.)
# Type in the R script or in the Console
x
#> [1] 1 2 3 1 3 25
y
#> [1] 5 31 71 1 3 21 6Objects exist in your R project workspace, listed in the RStudio Environment pane
Data are also named objects. For example, midfieldr has several toy data sets included for use in illustrative examples like this one. Type its name in the script,
# Examine a data frame included with midfieldr
toy_student
#> mcid institution transfer hours_transfer
#> <char> <char> <char> <num>
#> 1: MID25783939 Institution M First-Time in College NA
#> 2: MID25784402 Institution M First-Time in College NA
#> 3: MID25805538 Institution M First-Time in College NA
#> 4: MID25808099 Institution M First-Time in College NA
#> 5: MID25816437 Institution M First-Time in College NA
#> ---
#> 96: MID26656134 Institution L First-Time in College NA
#> 97: MID26656367 Institution L First-Time in College NA
#> 98: MID26663803 Institution L First-Time in College NA
#> 99: MID26678321 Institution L First-Time in College NA
#> 100: MID26692008 Institution L First-Time in College NA
#> race sex
#> <char> <char>
#> 1: White Female
#> 2: White Male
#> 3: White Female
#> 4: White Female
#> 5: White Male
#> ---
#> 96: Native American Male
#> 97: Hispanic/Latinx Male
#> 98: International Male
#> 99: White Female
#> 100: White MaleTo view the help page for the data, type in the Console
# type in the Console
? toy_studentIf we wanted the first five rows of the toy data, we use the [] operator.
# Practice using the `[` operator
toy_student[1:5]
#> mcid institution transfer hours_transfer race sex
#> <char> <char> <char> <num> <char> <char>
#> 1: MID25783939 Institution M First-Time in College NA White Female
#> 2: MID25784402 Institution M First-Time in College NA White Male
#> 3: MID25805538 Institution M First-Time in College NA White Female
#> 4: MID25808099 Institution M First-Time in College NA White Female
#> 5: MID25816437 Institution M First-Time in College NA White MaleTo view the help page for the [ operator, surround the symbol with “back-ticks” (on your keyboard with the tilde ~ symbol). For example,
# view the help page on the R extract operator
? `[`To extract a single column, e.g. the ID column, but preserve the data frame structure,
# Subset a column as a data table
toy_student[, .(mcid)]
#> mcid
#> <char>
#> 1: MID25783939
#> 2: MID25784402
#> 3: MID25805538
#> 4: MID25808099
#> 5: MID25816437
#> ---
#> 96: MID26656134
#> 97: MID26656367
#> 98: MID26663803
#> 99: MID26678321
#> 100: MID26692008We can also extract the column as a vector using slightly different syntax,
# Subset a column as a vector
toy_student[, mcid]
#> [1] "MID25783939" "MID25784402" "MID25805538" "MID25808099" "MID25816437"
#> [6] "MID25826223" "MID25828870" "MID25831839" "MID25839453" "MID25840802"
#> [11] "MID25841465" "MID25845841" "MID25846316" "MID25847220" "MID25848589"
#> [16] "MID25852023" "MID25853332" "MID25853799" "MID25877946" "MID25880643"
#> [21] "MID25887008" "MID25899243" "MID25911361" "MID25913454" "MID25931457"
#> [26] "MID25947836" "MID25982250" "MID25995980" "MID25997636" "MID26000057"
#> [31] "MID26004638" "MID26013461" "MID26020535" "MID26046521" "MID26048632"
#> [36] "MID26060301" "MID26062203" "MID26062778" "MID26086310" "MID26088450"
#> [41] "MID26102824" "MID26136319" "MID26138017" "MID26152744" "MID26161677"
#> [46] "MID26170598" "MID26173721" "MID26181209" "MID26187436" "MID26204281"
#> [51] "MID26211998" "MID26235812" "MID26244053" "MID26247839" "MID26305709"
#> [56] "MID26305863" "MID26309255" "MID26319252" "MID26332563" "MID26356320"
#> [61] "MID26358462" "MID26370377" "MID26383411" "MID26384771" "MID26391215"
#> [66] "MID26400804" "MID26413466" "MID26417039" "MID26418247" "MID26421588"
#> [71] "MID26421846" "MID26422829" "MID26429192" "MID26433811" "MID26435945"
#> [76] "MID26439623" "MID26441609" "MID26453554" "MID26461158" "MID26481120"
#> [81] "MID26526195" "MID26528318" "MID26546600" "MID26560837" "MID26561940"
#> [86] "MID26575282" "MID26577489" "MID26578111" "MID26588553" "MID26592425"
#> [91] "MID26592668" "MID26596818" "MID26605008" "MID26607528" "MID26655230"
#> [96] "MID26656134" "MID26656367" "MID26663803" "MID26678321" "MID26692008"Here you can see how the row labels in the printed output work. There are 5 IDs per row, so the second row starts with the 6th ID, indicated by [6]. The last row starts with the 96th value [96] and ends with the 100th value.
The “toy” data sets in midfieldr (toy_student, toy_course, toy_term, and toy_degree) include student unit records for only 100 students—not a statistically representative sample—used for package examples like those above.
5.5 R functions do things
Functions do something useful
- Functions are objects the perform actions for you
- Functions produce output based on the input it receives
- Functions are recognized by the parentheses at the end of their names
The parentheses are where we include the inputs (arguments) to the function
c()concatenates the comma-separated numbers in the parentheses to create a vectormean()computes the mean of a vector of numberssd()computes the standard deviation of a vector of numberssummary()returns a summary of the object
If we try mean() with no inputs, we get an error statement
mean()
#> Error in mean.default() : argument "x" is missing, with no defaultLet’s determine some summary statistics on our student transfer hours. Add these lines to your script, Save, and run.
# Extract a column as a vector
transfer_hours <- toy_student[, hours_transfer]
# Examine the vector
transfer_hours
#> [1] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA 30 55 NA 24 NA NA NA
#> [26] NA NA NA NA 4 NA 2 NA NA NA 1 7 1 3 1 5 NA NA NA NA NA NA NA NA NA
#> [51] NA NA NA NA NA NA NA NA NA NA 80 NA NA NA NA NA NA NA NA NA NA NA NA NA NA
#> [76] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA
# Operate on the vector
mean(transfer_hours)
#> [1] NAWe have to set the optional argument na.rm (“remove NA”) to take a mean
# Operate and ignore NA values
mean(transfer_hours, na.rm = TRUE)
#> [1] 17.75
# Another operation
sd(transfer_hours, na.rm = TRUE)
#> [1] 25.63068
# The summary also shows the count of NAs
summary(transfer_hours)
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 1.00 1.75 4.50 17.75 25.50 80.00 88Functions to examine a data frame.
names()Data frame column nameshead()andtail()First few and last few rows of a data frame
# Practice finding column names
names(toy_student)
#> [1] "mcid" "institution" "transfer" "hours_transfer"
#> [5] "race" "sex"
# Practice examining the first few rows of a data frame
head(toy_student)
#> mcid institution transfer hours_transfer race sex
#> <char> <char> <char> <num> <char> <char>
#> 1: MID25783939 Institution M First-Time in College NA White Female
#> 2: MID25784402 Institution M First-Time in College NA White Male
#> 3: MID25805538 Institution M First-Time in College NA White Female
#> 4: MID25808099 Institution M First-Time in College NA White Female
#> 5: MID25816437 Institution M First-Time in College NA White Male
#> 6: MID25826223 Institution M First-Time Transfer NA White Female
# Practice examining the last few rows of a data frame
tail(toy_student)
#> mcid institution transfer hours_transfer
#> <char> <char> <char> <num>
#> 1: MID26655230 Institution L First-Time in College NA
#> 2: MID26656134 Institution L First-Time in College NA
#> 3: MID26656367 Institution L First-Time in College NA
#> 4: MID26663803 Institution L First-Time in College NA
#> 5: MID26678321 Institution L First-Time in College NA
#> 6: MID26692008 Institution L First-Time in College NA
#> race sex
#> <char> <char>
#> 1: White Female
#> 2: Native American Male
#> 3: Hispanic/Latinx Male
#> 4: International Male
#> 5: White Female
#> 6: White MaleFunctions to examine columns (variables) in a data frame.
sort()andunique()often used togetheris.na()to return TRUE for every NA element in an object, otherwise FALSEsum()applied tois.na()converts logical TRUE to 1 and FALSE to 0 and adds the elements. The resulting integer is the number of NA values in the vector.
# Determine the unique values in a column
sort(unique(toy_student[, sex]))
#> [1] "Female" "Male"
# Find the rows with NA values.
is.na(toy_student[, sex])
#> [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [49] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [61] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [73] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [85] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [97] FALSE FALSE FALSE FALSE
# How many values are NA?
sum(is.na(toy_student[, sex]))
#> [1] 0Repeat for other columns.
# Practice examining another column
sort(unique(toy_student[, institution]))
#> [1] "Institution A" "Institution B" "Institution C" "Institution D"
#> [5] "Institution E" "Institution F" "Institution G" "Institution H"
#> [9] "Institution J" "Institution K" "Institution L" "Institution M"
sum(is.na(toy_student[, institution]))
#> [1] 0
# Practice examining another column
sort(unique(toy_student[, race]))
#> [1] "Asian" "Black" "Hispanic/Latinx" "International"
#> [5] "Native American" "Other/Unknown" "White"
sum(is.na(toy_student[, race]))
#> [1] 0
# Practice examining another column
sort(unique(toy_student[, hours_transfer]))
#> [1] 1 2 3 4 5 7 24 30 55 80
sum(is.na(toy_student[, hours_transfer]))
#> [1] 88The help pages for functions are accessed via the Console. By viewing the help page you can find descriptions of arguments and their default settings if any. Try a few:
? mean()? sd()? summary()? names()? head()? sort()? unique()? is.na()? sum()
5.6 R functions come in packages
Functions are bundled in packages
- Families of useful functions are bundled into packages that you can install, load, and use
- Packages allow you to build on the work of others
- You can write your own functions and packages too
- A lot of the work in data science consists of choosing the right functions and giving them the right arguments to get our data into the form we need for analysis or visualization
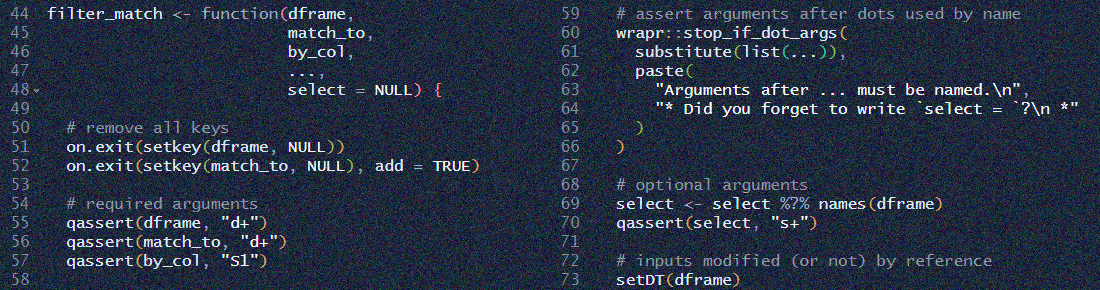
For example, to see the list of functions in the midfieldr package,
# Showing all functions in the midfieldr package
sort(getNamespaceExports("midfieldr"))
#> [1] "add_completion_timely" "add_data_sufficiency" "add_institution"
#> [4] "add_race_sex" "add_timely_term" "condition_fye"
#> [7] "condition_multiway" "filter_match" "filter_search"To view a help page, type, e.g.,
# Type in the Console to view a help page
? add_race_sex() In contrast, do the same for the data.table package,
# A package with many functions.
sort(getNamespaceExports("data.table"))
#> [1] "%between%" "%chin%" "%flike%"
#> [4] "%ilike%" "%inrange%" "%like%"
#> [7] ".__C__data.table" ".__C__IDate" ".__C__ITime"
#> [10] ".__T__$:base" ".__T__$<-:base" ".__T__[:base"
#> [13] ".__T__[[<-:base" ".__T__[<-:base" ".BY"
#> [16] ".EACHI" ".GRP" ".I"
#> [19] ".Last.updated" ".N" ".NGRP"
#> [22] ".rbind.data.table" ".SD" ":="
#> [25] "address" "alloc.col" "as.data.table"
#> [28] "as.IDate" "as.ITime" "as.xts.data.table"
#> [31] "between" "chgroup" "chmatch"
#> [34] "chorder" "CJ" "copy"
#> [37] "cube" "data.table" "dcast"
#> [40] "dcast.data.table" "fcase" "fcoalesce"
#> [43] "fifelse" "fintersect" "first"
#> ... etc. truncatedDon’t panic! We will use only a small number of these functions from data.table. For example, the %ilike% function, view its help page by running
# Type in the Console to view the help page
? `%ilike%` 5.7 R objects have class
Everything is an object and every object has a class.
class(x)
#> [1] "numeric"
class(summary)
#> [1] "function"Certain actions will change the class of an object. Suppose we try create a vector from the x object and a text string,
new_vector <- c(x, "Apple")
new_vector
#> [1] "1" "2" "3" "1" "3" "25" "Apple"
class(new_vector)
#> [1] "character"By adding the word “Apple” to the vector, R changed the class from “numeric” to “character.” All the numbers are enclosed in quotes: they are now character strings and cannot be used in calculations.
The most common class of data object we will use is the data frame. The data in midfieldr are stored as data frames, e.g.,
# examine another midfieldr data set
study_stickiness
#> program race sex ever grad stick
#> <char> <char> <char> <int> <int> <num>
#> 1: Civil Asian Female 17 12 70.6
#> 2: Civil Black Female 54 28 51.9
#> 3: Civil White Female 329 232 70.5
#> 4: Civil Asian Male 37 24 64.9
#> 5: Civil Black Male 98 43 43.9
#> ---
#> 34: Mechanical Hispanic/Latinx Male 76 47 61.8
#> 35: Mechanical International Male 37 19 51.4
#> 36: Mechanical Native American Male 14 8 57.1
#> 37: Mechanical Other/Unknown Male 48 28 58.3
#> 38: Mechanical White Male 1940 1265 65.2
class(study_stickiness)
#> [1] "data.table" "data.frame"- Six columns: program, race, sex, ever, grad, stick.
- Three columns are labeled
<char>for character, categorical variables
- Two columns are labeled
<int>for integer - One column is labeled
<num>for double precision
The additional class shown data.table is an augmented version of the base R data.frame class. When working with these objects you can use base R data.frame syntax or data.table syntax.
If you have a data.frame object that is not a data.table, e.g. the airquality data frame that comes with R
class(airquality)
#> [1] "data.frame"
head(airquality)
#> Ozone Solar.R Wind Temp Month Day
#> 1 41 190 7.4 67 5 1
#> 2 36 118 8.0 72 5 2
#> 3 12 149 12.6 74 5 3
#> 4 18 313 11.5 62 5 4
#> 5 NA NA 14.3 56 5 5
#> 6 28 NA 14.9 66 5 6You can convert it to data.table object with as.data.table() and assign it to a slightly different name.
air_quality <- as.data.table(airquality)
class(air_quality)
#> [1] "data.table" "data.frame"
air_quality
#> Ozone Solar.R Wind Temp Month Day
#> <int> <int> <num> <int> <int> <int>
#> 1: 41 190 7.4 67 5 1
#> 2: 36 118 8.0 72 5 2
#> 3: 12 149 12.6 74 5 3
#> 4: 18 313 11.5 62 5 4
#> 5: NA NA 14.3 56 5 5
#> ---
#> 149: 30 193 6.9 70 9 26
#> 150: NA 145 13.2 77 9 27
#> 151: 14 191 14.3 75 9 28
#> 152: 18 131 8.0 76 9 29
#> 153: 20 223 11.5 68 9 30The data frame as a whole has a class; so do the individual columns.
class(air_quality[, Ozone])
#> [1] "integer"
class(air_quality)
#> [1] "data.table" "data.frame"5.8 R objects have structure
To see inside an object ask for its structure using the str() function.
str(x)
#> num [1:6] 1 2 3 1 3 25
str(toy_term)
#> Classes 'data.table' and 'data.frame': 169 obs. of 6 variables:
#> $ mcid : chr "MID25899243" "MID26319252" "MID25841465" "MID26560837" ...
#> $ institution: chr "Institution B" "Institution E" "Institution M" "Institution J" ...
#> $ term : chr "19943" "20021" "20023" "19981" ...
#> $ cip6 : chr "240102" "140801" "260101" "999999" ...
#> $ level : chr "03 Junior" "04 Senior" "02 Sophomore" "02 Sophomore" ...
#> $ hours_term : num 5 5 16 12 15 21 9 12 17 17 ...
#> - attr(*, ".internal.selfref")=<externalptr>
str(airquality)
#> 'data.frame': 153 obs. of 6 variables:
#> $ Ozone : int 41 36 12 18 NA 28 23 19 8 NA ...
#> $ Solar.R: int 190 118 149 313 NA NA 299 99 19 194 ...
#> $ Wind : num 7.4 8 12.6 11.5 14.3 14.9 8.6 13.8 20.1 8.6 ...
#> $ Temp : int 67 72 74 62 56 66 65 59 61 69 ...
#> $ Month : int 5 5 5 5 5 5 5 5 5 5 ...
#> $ Day : int 1 2 3 4 5 6 7 8 9 10 ...
str(air_quality)
#> Classes 'data.table' and 'data.frame': 153 obs. of 6 variables:
#> $ Ozone : int 41 36 12 18 NA 28 23 19 8 NA ...
#> $ Solar.R: int 190 118 149 313 NA NA 299 99 19 194 ...
#> $ Wind : num 7.4 8 12.6 11.5 14.3 14.9 8.6 13.8 20.1 8.6 ...
#> $ Temp : int 67 72 74 62 56 66 65 59 61 69 ...
#> $ Month : int 5 5 5 5 5 5 5 5 5 5 ...
#> $ Day : int 1 2 3 4 5 6 7 8 9 10 ...
#> - attr(*, ".internal.selfref")=<externalptr>5.9 R syntax comes in many flavors
As you work through the MIDFIELD tutorials, we’ll introduce syntax by example from both base R and the data.table package. There can be subtle differences that we will attempt to avoid by consistent usage.
For example, to subset a column from a data frame but keep it as a column (not a vector), the base R syntax is
# base R subset one column
airquality[, "Ozone", drop = FALSE]
#> Ozone
#> 1 41
#> 2 36
#> 3 12
#> 4 18
#> 5 NA
#> 6 28
#> 7 23
#> 8 19
#> 9 8
#> 10 NA
#> ..., etc., truncated for brevityIn data.table syntax, the same operation is as follows. Note we are using the air_quality data.table object we created earlier.
# data.table extract one column
air_quality[, .(Ozone)]
#> Ozone
#> <int>
#> 1: 41
#> 2: 36
#> 3: 12
#> 4: 18
#> 5: NA
#> ---
#> 149: 30
#> 150: NA
#> 151: 14
#> 152: 18
#> 153: 20If we want the same information extracted as a vector, we would:
# base R subset one column as a vector
airquality[, "Ozone"]
#> [1] 41 36 12 18 NA 28 23 19 8 NA 7 16 11 14 18 14 34 6
#> [19] 30 11 1 11 4 32 NA NA NA 23 45 115 37 NA NA NA NA NA
#> [37] NA 29 NA 71 39 NA NA 23 NA NA 21 37 20 12 13 NA NA NA
#> [55] NA NA NA NA NA NA NA 135 49 32 NA 64 40 77 97 97 85 NA
#> [73] 10 27 NA 7 48 35 61 79 63 16 NA NA 80 108 20 52 82 50
#> [91] 64 59 39 9 16 78 35 66 122 89 110 NA NA 44 28 65 NA 22
#> [109] 59 23 31 44 21 9 NA 45 168 73 NA 76 118 84 85 96 78 73
#> [127] 91 47 32 20 23 21 24 44 21 28 9 13 46 18 13 24 16 13
#> [145] 23 36 7 14 30 NA 14 18 20
# data.table subset one column as a vector
air_quality[, Ozone]
#> [1] 41 36 12 18 NA 28 23 19 8 NA 7 16 11 14 18 14 34 6
#> [19] 30 11 1 11 4 32 NA NA NA 23 45 115 37 NA NA NA NA NA
#> [37] NA 29 NA 71 39 NA NA 23 NA NA 21 37 20 12 13 NA NA NA
#> [55] NA NA NA NA NA NA NA 135 49 32 NA 64 40 77 97 97 85 NA
#> [73] 10 27 NA 7 48 35 61 79 63 16 NA NA 80 108 20 52 82 50
#> [91] 64 59 39 9 16 78 35 66 122 89 110 NA NA 44 28 65 NA 22
#> [109] 59 23 31 44 21 9 NA 45 168 73 NA 76 118 84 85 96 78 73
#> [127] 91 47 32 20 23 21 24 44 21 28 9 13 46 18 13 24 16 13
#> [145] 23 36 7 14 30 NA 14 18 20In each case, you will note that the square bracket operators include a comma [i, j]. In general, the comma separates row operations i from column operations j. When there is no row operation (as in the examples above), the i position is empty.
In data table syntax, we go one step further and add a grouping index, that is
DT[i, j, by]This can be read as “Take DT, subset/reorder rows using i, then calculate j, grouped by by.” The midfieldr tutorials illustrate this usage in practice. For more detail you can work through the Data basics tutorial.
5.10 Help with R
Online resources
Tutorial
- Getting Started in R: Tinyverse Edition. Highly recommended. An 8-page introduction to R using data.table and ggplot2.
Online Q & A
- Stack Overfow R section. A question-and-answer site.
Cheat sheets. Compact (information dense) summaries of features.
Package main help
For a package’s main help page, help(package = "name_of_package"), to obtain a list of all the functions (and possibly some data objects) in the package.
# type in the Console
help(package = "midfieldr")In the Help pane, click through any of the links for details on the function.
Package vignettes
Some packages come with vignettes, articles explaining how to use the package for specific tasks. For example,
# type in the Console
browseVignettes(package = "data.table")
browseVignettes(package = "ggplot2")
# to see a listing of all vignettes in all your installed packages
browseVignettes()Function help
A number of times so far we’ve shown how to get the help page for a function when we know its name, e.g.,
# type in the Console
? str()If you recall only a part of the function name, use apropos() to search packages that are currently loaded. “Loading” a package is what we do when we use the library() function at the top of a script.
For example, midfieldr has functions with “timely” in their name because they involve timely completion. apropos() reports two functions:
# type in the Console
apropos("timely")
#> [1] "add_completion_timely" "add_timely_term"Examples
There are often examples at the end of an R function help page. You can run them using the example() function. For example,
# base R function examples
example("mean")
#>
#> mean> x <- c(0:10, 50)
#>
#> mean> xm <- mean(x)
#>
#> mean> c(xm, mean(x, trim = 0.10))
#> [1] 8.75 5.50try one of the midfieldr function examples,
# midfieldr function examples
example("add_institution")
#>
#> add_ns> # Extract a column of IDs from student
#> add_ns> id <- toy_student[, .(mcid)]
#>
#> add_ns> # Add institutions from term
#> add_ns> DT1 <- add_institution(id, midfield_term = toy_term)
#>
#> add_ns> head(DT1)
#> mcid institution
#> <char> <char>
#> 1: MID25783939 Institution M
#> 2: MID25784402 Institution M
#> 3: MID25805538 Institution M
#> 4: MID25808099 Institution M
#> 5: MID25816437 Institution M
#> 6: MID25826223 Institution M
#>
#> add_ns> nrow(DT1)
#> [1] 100
#>
#> add_ns> # Will overwrite institution column if present
#> add_ns> DT2 <- add_institution(DT1, midfield_term = toy_term)
#>
#> add_ns> head(DT2)
#> mcid institution
#> <char> <char>
#> 1: MID25783939 Institution M
#> 2: MID25784402 Institution M
#> 3: MID25805538 Institution M
#> 4: MID25808099 Institution M
#> 5: MID25816437 Institution M
#> 6: MID25826223 Institution M
#>
#> add_ns> nrow(DT2)
#> [1] 1005.11 Keyboard shortcuts
If you are working in RStudio, you can see the menu of keyboard shortcuts using the menu Tools > Keyboard Shortcuts Help. The shortcuts we use regularly include
| Windows / Linux | Action | Mac OS |
|---|---|---|
ctrl L |
Clear the RStudio Console | ctrl L |
ctrl shift C |
Comment/uncomment line(s) | cmd shift C |
ctrl X, C, V |
Cut, copy, paste | cmd X, C, V |
ctrl F |
Find in text | cmd F |
ctrl I |
Indent or re-indent lines | cmd I |
alt \(-\) |
Insert the assignment operator <- |
option \(-\) |
ctrl alt B |
Run from beginning to line | cmd option B |
ctrl alt E |
Run from line to end | cmd option E |
ctrl Enter |
Run selected line(s) | cmd Return |
ctrl S |
Save | cmd S |
ctrl A |
Select all text | cmd A |
ctrl Z |
Undo | cmd Z |
Lastly, any time you want a fresh start in your working environment,
- Use the pulldown menu Session > Restart R
5.12 Next steps
That concludes our brief introduction to R basics. Select one of the links to continue your progression.